
- MASS FINDER SCIENCE SOFTWARE
- MASS FINDER SCIENCE WINDOWS 7
If you want to perform the MS/MS peak annotation with the known structure, prepare two fields including FORMULA and SMILES.Mass spectrum is supposed to be imported as MS/MS.M/z intensity pair (tab, comma, space can be used as the delimiter.) The format of MSP basically follows the NIST MS search manual. Unknown queries should be separately stored in the ASCII file: the MSP or MAP file ‘CANNOT’ store multi compound records in the single file. MSP or MAT formatted by the following explanations. This program accepts two file extensions, i.e. Moreover, this program can be called from the MS-DIAL program which is downloadable at. In addition, the users can directly make the query in the MS-FINDER graphical user interface. The program can import ASCII format files including MSP (EI-MS and MS/MS) or improved MSP (both MS and MS/MS, the file extension must be MAT.).
MASS FINDER SCIENCE SOFTWARE
MS-FINDER can be used as the local software program in Windows PC.
MASS FINDER SCIENCE WINDOWS 7
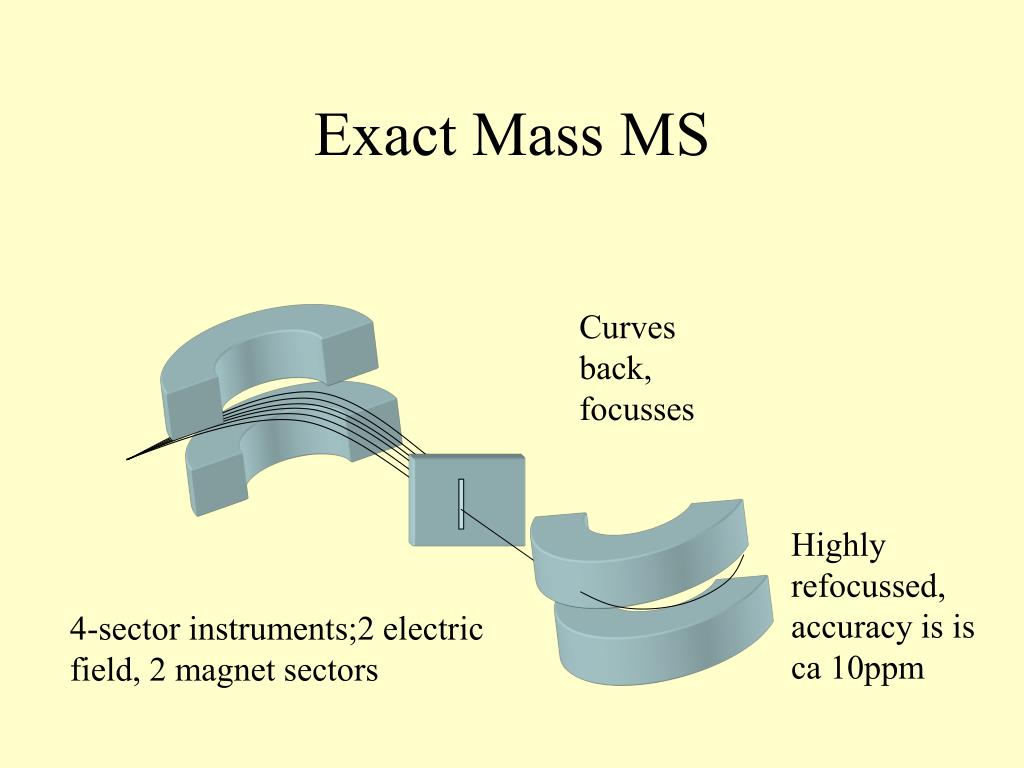
Windows OS (.NET Framework 4.5 or later): Windows 7 or later. Section 12: Molecular spectrum networking Section 9: Compound annotation (batch analysis) Section 8: FSEA: fragment set enrichment analysis Section 7: Compound annotation by searching spectral databases Section 6: Compound annotation by in silico fragmenter Section 4-2: From the graphical user interface of the MS-FINDER program Section 4-1: From a folder which includes MSP or MAT format files Section 3-6: Specific field to fix the formula element count Section 3-5: User defined structure database format Section 3-4: Adduct ion format: +, 2+, +, -, etc. Oliver Fiehn team (UC Davis) supported by the JST/NSF SICORP ‘Metabolomics for the low carbon society’ project. Masanori Arita team (RIKEN, Reifycs Inc.) and Prof. MS-FINDER has been developed as the collaborative work between Prof. In addition, the program can annotate your unknowns by the public spectral databases such as MassBank, LipidBlast, and GNPS. First, MS-FINDER aims to provide solutions for 1) formula predictions, 2) fragment annotations, and 3) structure elucidations by means of unknown spectra. MS-FINDER was launched as a universal program for compound ‘annotation’ that supports EI-MS (GC/MS) and MS/MS spectral mining. Currently, the main bottleneck of GC/MS- and LC/MS/MS based untargeted analysis is compound identification due to the limitation of EI-MS and MS/MS records of authentic standard. Gas chromatography coupled with electron ionization mass spectrometer (GC/MS) and liquid chromatography coupled with electrospray ionization- (ESI-) tandem mass spectrometer (LC/MS/MS) are the preferred tools for untargeted metabolomics. The purpose of metabolomics is to perform the ‘comprehensive’ analysis for small biomolecules of living organisms.





 0 kommentar(er)
0 kommentar(er)
